International Journal of Epidemiology And Public Health Research
OPEN ACCESS | Volume 9 - Issue 1 - 2026
ISSN No: 2836-2810 | Journal DOI: 10.61148/2836-2810/IJEPHR
Anukumar B1 and Sugunan A P2
1Scientist & Officer in charge, ICMR-National Institute of Virology, Alappuzha, Kerala.
2Scientist, ICMR-National Institute of Virology, Alappuzha, Kerala.
*Corresponding author: Anukumar B, Scientist & Officer in charge, ICMR-National Institute of Virology, Alappuzha, Kerala.
Received: May 01, 2025
Accepted: May 09, 2025
Published: May 12, 2025
Citation: Anukumar B and Sugunan A P. (2025) “An outbreak of norovirus associated acute gastroenteritis in the Southern District of Kerala”. International Journal of Epidemiology and Public Health Research, 6(3); DOI: 10.61148/2836- 2810/IJEPHR/126.
Copyright: © 2025. Anukumar B. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited., provided the original work is properly cited.
A widespread acute gastroenteritis outbreak was reported in Alappuzha, Kerala, during the third week of June 2021. Symptoms such as vomiting and diarrhea have been observed in people in all age groups. There were no fatalities among those who were infected. A descriptive study performed to determine the epidemiological characteristics and etiology of the outbreak. A realstar Norovirus RT‒PCR kit was used to detect norovirus in the stool samples. A partial junction sequence of ORF1/ORF-2 was used for sequence analysis. A total of 770 cases of acute gastroenteritis from the same area were reported at a public health facility from June‒July 2021. Among the reported cases, children under 5 years of age were the most affected, with an attack rate of 10.3. Six out of the eleven stool samples referred to the National Institute of Virology, Kerala unit were positive for norovirus GII. The phylogenetic tree revealed that the Kerala strain is closely related to the Japan 2015 strain and has 99% homology with GII.P17 type is a widely circulating strain in other countries. The study concluded that the outbreak was caused by norovirus GII. The virus may circulate locally in India and may be introduced into Kerala for the first time
Introduction:
Acute gastroenteritis (AGE) is one of the most common diseases in developing countries (1). Human norovirus (NoV), a positive sense, single-stranded RNA (7.6 kb) virus of the Caliciviridae family, is a major causative agent of food- and water-borne outbreaks of nonbacterial acute gastroenteritis. The genome has three open reading frames (ORFs). ORF1 encodes six nonstructural proteins and ORF2 and ORF3 encode major and minor capsid proteins, respectively. NoVs are classified into 10 genogroups (GI-GX) and more than 45 genotypes (2). Among these, genogroups I, II, and IV are reportedly associated with human infections (3). Genogroup II (GII) is the predominant genogroup, accounting for more than 70% of NoV outbreaks. Nagamani et al., reported the emergence of a new variant viz., GII.4, in India (4). NoV affects all age groups of people irrespective of their socioeconomic setting. NoV has a low infectious dose (18 to 1,000 viral particles), high stability (0‒60oC), is a subclinical or healthy carrier, is highly diverse, has frequent genomic mutations (via antigenic shifting and recombination) (5) and induces only very short- term immunity. Owing to these features, they are well known for their rapid spread in closed (school/hostel canteens, cruise ships, military camps) or overcrowded communities. During the past 6 years, hundreds of food- and water-borne outbreaks have been reported by the Integrated Disease Surveillance Program (IDSP). NoV is well known for its transmission via ground water. Symptoms of NoV include diarrhea, vomiting, nausea, stomach pain, fever, headache and body aches, although NoV infections can also be asymptomatic. The transmission of NoV occurs primarily via direct person‒person contact, fecal‒oral route, and the consumption of contaminated food or water (6).
In June 2021, an outbreak of gastroenteritis was reported in the municipality of Alappuzha in Kerala. Approximately 770 cases were reported to the IDSP by mid-July 2021, with no causality. The area has more than 15000 dwelling houses with 2 lakhs people. We investigated the outbreak to describe its epidemiological characteristics and to determine its etiology.
Methods:
Ethics statement: The study was approved by the ICMR-National Institute of Virology, Institute Human Ethical Committee (Approval No. NIV/IEC/Mar/2022/D-22).
Case definition
The IDSP’s case definition of AGE, which is any patient reporting the passage of ≥3 watery stools within 24 hours and vomiting, was used to identify cases. Data on AGE cases reported from Alappuzha town and its suburbs by the district IDSP from 21st June to 15th July, 2021were obtained. A house-to-house survey was conducted by the ICMR-National Institute of Virology Kerala Unit (NIV-Kerala unit) team involving an epidemiologist and clinician, a microbiologist, and data entry staff to obtain epidemiological and clinical details from a subsample of the cases in that area. A regionally modified WHO questionnaire on acute diarrheal disease was used to collect epidemiological details (7). Approximately 423 cases were contacted via telephone, when house visits were not possible. Data on the population at risk were obtained from census publications, and the current population was estimated via extrapolation via an exponential growth model (Census of India 2011DDW_PCA0000_2011_Indiastatedist.xlsx)
Laboratory diagnosis
Aliquots of stool/rectal swab samples from 11 suspected cases were collected at the NIV-Kerala unit from the respective government health facility to investigate the viral etiology. Other aliquots of the same samples were processed for bacterial enteric pathogens in the Microbiology Department of Govt.T.D.Medical College, Alappuzha. The samples were tested for rotavirus and enterovirus by in-house RT‒PCR and norovirus by Real®Star Norovirus RT‒PCR kit (Altona Diagnostics GmbH, Germany).
Norovirus sequence analyses
Primers covering the overlapping region of the ORF1/ORF2 genes (584bp) including the junction sequences of norovirus, were synthesised. High-fidelity Phusion polymerase was used to amplify fragments of approximately 600 bp (Thermo Fisher, USA). A NucleoSpin Gel and PCR clean-up kit (Macherey-Nagel, Germany) was used to gel purify the amplified PCR products. Gene sequencing of the purified amplicons was carried out at a commercial gene sequencing agency (AgriGenome Labs Pvt.Ltd, India).
Phylogenetic analysis
Phylogenetic analysis was performed in MEGA 7.0. Genome sequences (n=17) available in GenBank representing recent outbreaks were retrieved for analysis. The 17 sequences consisted of sequences from India and sequences from other countries. The sequences obtained from GenBank were aligned via ClustalW mode in MEGA v 7.0.26 software (8). A phylogenetic tree was constructed via the maximum likelihood method in MEGA. Genetic distances were calculated via the Tamura Nei model of nucleotide substitution. The robustness of the resulting tree was assessed with 1000 bootstrap replicates.
Statistical analysis
An epidemic curve was drawn for reported cases until the end of July-2021, and age and sex wise attack rates were calculated based on the data obtained via a house-to-house survey.
Results:
A total of 770 AGE cases (52% males) were reported by the IDSP from June‒July 2021. A cluster of 56 cases during the last week of June represented the beginning of the outbreak. By the 2nd week of July, approximately 450 similar cases were reported without any mortality. The total population in the area was 215,708, and the outbreak was confirmed by comparing the baseline incidence of AGE in Alappuzha in previous years by the IDSP. The epidemic curve showed an initial peak on June 27, 2021, and three secondary peaks subsequently occurred (Fig.1). By the second week of July, the number of cases had decreased rapidly, and the outbreak had subsided.
The most common symptom was vomiting (75%) followed by diarrhea and malaise (60% and 50%, respectively). Few of them reported having fever or abdominal pain. In this study, the attack rate among males (2.9 per 1000 people) was greater than that among females (1.9 per 1000 people). Most of the age groups affected was <5 years-old children (33%), with an attack rate of 10.3 per 1000 people. Owing to fast recovery and fear of wage losses, many unreported cases were also present. No history of common events was noted among the cases other than the routine consumption of municipal water contaminated with ground water.
The common bacterial pathogens causing AGE were ruled out at a government laboratory. At NIV, the stool samples/rectal swabs were negative for rotavirus and enterovirus. Among the 11 samples, six (54%) were positive for norovirus GII. Basic Local Alignment Search Tool (BLAST, GenBank, NCBI) analysis of 584 nt sequences derived from the ORF1/ORF2 region (Acc.No. ON629765) showed that the Kerala strain is very close to the GII P.17 type and has 99% identity with the 2015 strain from Japan (Fig.2). This study confirmed that the outbreak was caused by norovirus GII.
Discussion:
NoV is the major cause of acute gastroenteritis related to food and waterborne outbreaks, and sporadic cases have appeared worldwide, especially in developing countries. In our study, we confirmed the etiology as norovirus GII, and children were affected more among the cases. This virus was reported for the first time in Kerala. Following this outbreak in Alappuzha, several sporadic outbreaks have occurred in different districts of Kerala. The most common symptom present in our study was vomiting (75%), whereas Sai L et al., reported that 67.5% of children experienced vomiting and that 46.3% experienced fever (9). In contrast, a study from Western India reported that 32% of hospitalized outpatients and 43% of outpatients did not experience vomiting (10). Another study reported the absence of vomiting in 51% of norovirus-infected patients (11). Although norovirus was detected in all age groups, infection was more common in children less than 5 years of age (33%). This observation was in accordance with previous studies (10,12). Person-to-person transmission may have played an important role in this outbreak. People living in the same area usually have frequent contact during gatherings and other activities. Airborne or fomite transmission may also facilitate the spread of the virus during outbreaks (13). The defined study area shares a common water source contaminated with stagnant water. The probable source of infection could be stagnant water. The source of infection could not be identified from suspected samples because of resource constraints.
A phylogenetic study revealed that the strain is very close to the 2015 Japanese strain. NCBI BLAST analysis revealed that the Kerala strain is 99% similar to GII.P17 genotype. GII.P17 was first reported in India. However, complete RdRp gene sequencing is necessary to determine the P group of noroviruses GII. The genotype emerged during the period of 2014‒2015 in East Asian countries (14‒15), and the genotype slowly replaced the predominant GII.4 genotype (16‒17). Although the disease is called winter vomiting disease in the western part of the world, many reports have shown that the disease also occurs in the summer season (10, 18).
Conclusion:
Unidentified acute gastroenteritis cases need to be ruled out for NoV thus identifying the potential threat of environmental contamination, although we could not perform environmental sample testing. Public concerns regarding NoV have recently increased worldwide. Hence, the system of transmission of various NoVs needs to be studied molecularly as well as epidemiologically. Such enhanced knowledge might provide salient evidence for vaccine development along with the detection of new emerging variants of NoV via comparisons.
Acknowledgement:
We gratefully acknowledge the director, NIV, for support of the study. We also acknowledge the Department of Health Research for funding the study through the Viral Research and Diagnostic Laboratories Network program. We thank the District Medical Officer, Alappuzha, the outbreak investigation team of ICMR-National Institute of Epidemiology, NIV Kerala unit staff and the NIV Manuscript Review Committee.
Conflict of interest: The authors declare no conflicts of interest for this article
Author contribution: Dr. Sugunan analyzed the data and reviewed the manuscript, whereas Dr. Anukumar performed the genetic analysis and reviewed the manuscript.
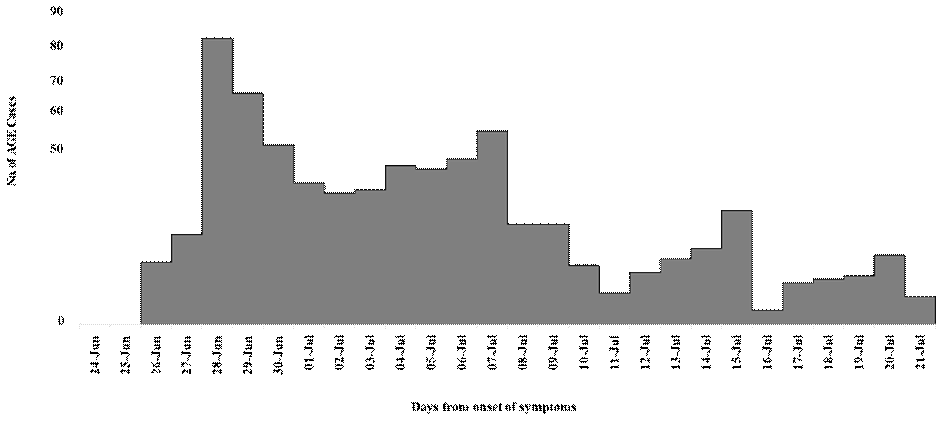
Figure 1: Epidemic curve of acute gastroenteritis outbreak in Alappuzha Municipality, Kerala
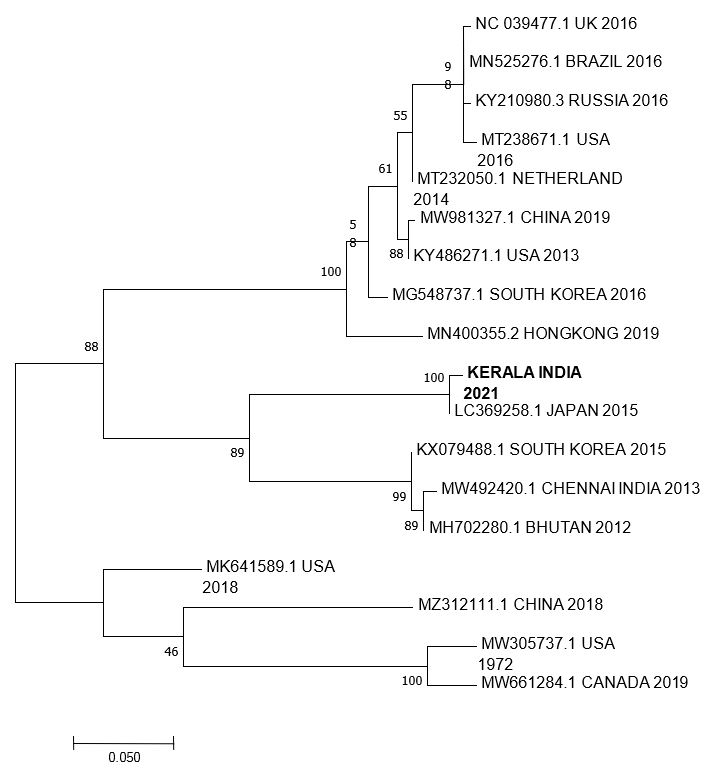
Figure 2: Phylogenetic tree of Norovirus from Kerala and reference sequences from GenBank. Sequences were aligned using CLUSTALW (1000 bootstrap replicates), and phylogenetic inferences were obtained using the maximum-likelihood method within the MEGA7. The scale bar corresponds to 0.05 change per nucleotide. Sequence from Kerala is indicated by bold letters, and reference sequences are indicated by accession number and region.