Clinical Case Reports and Clinical Study
OPEN ACCESS | Volume 13 - Issue 1 - 2026
ISSN No: 2766-8614 | Journal DOI: 10.61148/2766-8614/JCCRCS
Sorush Niknamian
MD PHD MPH Dr.PH, Military Medicine Dep., Liberty University, USA
Fellow Member of International Society of Infectious Diseases (ISID)
Member of Department of Defense (DoD), Military, Military Medicine Dep., USA Member of UN and MSF
*Corresponding author: Sorush Niknamian, Military Medicine Dep., Liberty University, USA
Received: February 19, 2021
Accepted: February 26, 2021
Published: March 06, 2021
Citation: Niknamian S. “ The Negative Impact of The Hominin’s Dpp4 Gene Inherited from Neanderthals To Pandemic of Covid-19”. Clinical Case Reports and Clinical Study, 3(1) ; DOI: 10.61148/2766-8614/JCCRCS/033
Copyright: © 2021 Sorush Niknamian. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Background: According to preliminary sequences from 2010, 99.7% of the nucleotide sequences of the modern human and Neanderthal genomes are identical, compared to humans sharing around 98.8% of sequences with the chimpanzee. ... In contrast, the difference between chimpanzees and modern humans is approximately 1,462 mtDNA base pairs.
Materials and Methods: Neanderthal-inherited genetic material is found in all non-African populations and was initially reported to comprise 1 to 4 percent of the genome. This fraction was later refined to 1.5 to 2.1 percent. We had gone through many researches of Neanderthals affected gene flow in humans.
Results: It is estimated that 20 percent of Neanderthal DNA currently survives in modern humans. Modern human genes involved in making keratin, a protein constituent of skin, hair, and nails, have especially high levels of introgression. For example, approximately 66% of East Asians contain a POUF23L variant introgressed from Neanderthals, while 70% of Europeans possess an introgressed allele of BNC2. Our finding shines a light on an enzyme called dipeptidyl peptidase-4 (DPP4). Scientists already know the protein allows another coronavirus, which causes Middle Eastern respiratory syndrome (MERS), to bind to and enter human cells. The new analysis, of DPP4 gene variants among COVID-19 patients, suggests the enzyme also provides SARS-CoV-2 with a second door into our cells, along with its usual infection route via the angiotensin-converting enzyme 2 (ACE2) receptor on cell surfaces. Conclusion: Most Europeans, Asians, and Native Americans harbor a handful of genes from Neanderthals, up 1.8% to 2.6% of their DNA. Studies of ancient DNA in Neanderthal fossils have shown the hominin's DPP4 gene subtly differs from the typical human one.
Conclusion: The hominin's DPP4 gene inherited from Neanderthals plays a major role in Immune System Disorders and Lower Immune response in many diseases. This gene plays a major role in affecting humans with COVID-19 and spreading it through the world. All humans contain this gene from 1 to 4 percent. East Asians, Europeans, Middle and south Americans conveys more, hence; native Africans contain less amounts of hominin's DPP4 gene. Therefore; East Asians, Europeans, Middle and south Americans are prone to severe COVID-19.
Introduction
Neanderthal ancient DNA
Genetic studies on Neanderthal ancient DNA became possible in the late 1990s. [2] The Neanderthal genome project, established in 2006, presented the first fully sequenced Neanderthal genome in 2013. Since 2005, evidence for the substantial admixture of Neanderthals DNA in modern populations has accumulated. [3] The divergence time between the Neanderthal and modern human lineages is estimated at between 750,000 and 400,000 years ago. The more recent time depth has been suggested by Endicott et al. (2010) [4] and Rieux et al. (2014) [5] A significantly deeper time of separation, combined with repeated early admixture events, was calculated by Rogers et al. (2017). [6]. On July 3, 2020, a team reported finding that a major genetic risk factor of the Covid-19 virus was inherited from archaic Neanderthals 60,000 years ago. [7-8]
COVID-19
SARS-CoV-19 which is better called COVID-21, are a group of viruses that cause diseases in mammals and birds. In humans, the beginning of Coronaviruses in 1919 causes respiratory tract infections that are typically mild, such as some cases of the common cold. Rarer forms can be lethal, such as SARS, MERS, and COVID-19. Symptoms vary in other species: in chickens, they cause an upper respiratory tract disease, while in cows and pigs they cause diarrhea. Coronaviruses constitute the subfamily Orthocoronavirinae, The genome size, coronaviruses ranges from approximately 27 to 34 kilobases, the largest among known RNA viruses. In recent months, COVID-19 has become more severe which targets Respiratory Organ, Liver, Kidneys, Hearts and etc. The polarity of this virus is positive-sense ((+) ssRNA). Positive sense viral RNA is similar to mRNA and thus can be immediately translated by the host cell. Recombination in RNA viruses appears to be an adaptation for coping with genome damage. Recombination can occur infrequently between animal viruses of the same species but of divergent lineages. The resulting recombinant viruses may sometimes cause an outbreak of infection in humans. RNA viruses have very high mutation rates This is one reason why it is difficult to make effective vaccines to prevent diseases caused by RNA viruses. The resulting recombinant viruses causes an outbreak of infection in humans. [Sorush Niknamian et al. 2021]
Materials and Methods
The question of possible interbreeding between Neanderthals and anatomically modern humans (AMH) had been looked into since the early archaeogenetic studies of the 1990s. In 2006, no evidence for interbreeding had yet been found. [9] In 2009, analysis of about one-third of the full genome of the Altai individual was still reported as showing no sign of admixture. The variant of microcephalin common outside Africa, which was suggested to be of Neanderthal origin and responsible for rapid brain growth in humans, was not found in Neanderthals. Nor was a very old MAPT variant which is found primarily in Europeans. [10] Positive evidence for admixture was first published in May 2010. [11] Neanderthal-inherited genetic material is found in all non-African populations and was initially reported to comprise 1 to 4 percent of the genome. [12] This fraction was later refined to 1.5 to 2.1 percent. [13]
It is estimated that 20 percent of Neanderthal DNA currently survives in modern humans. [14] Modern human genes involved in making keratin, a protein constituent of skin, hair, and nails, have especially high levels of introgression. For example, approximately 66% of East Asians contain a POUF23L variant introgressed from Neanderthals, while 70% of Europeans possess an introgressed allele of BNC2.
Neanderthal variants affect the risk of developing several diseases, including lupus, biliary cirrhosis, Crohn's disease, type 2 diabetes, and severe COVID-19. [15-17] The allele of MC1R which was originally linked to red hair in Neanderthals is not found in Europeans, but is present in Taiwanese Aborigines at a frequency of 70% and moderately high frequencies in other East Asian populations; hence, there is no evidence that Neanderthals had red hair.[18] While interbreeding is viewed as the most parsimonious interpretation of these genetic findings, the 2010 study still could not conclusively rule out an alternative scenario, in which the source population of non-African modern humans was already more closely related to Neanderthals than other Africans were, because of ancient genetic divisions within Africa.[19] [20] Research since 2010 has refined the picture of interbreeding between Neanderthals, Denisovans, and anatomically modern humans. Interbreeding appears to have occurred asymmetrically among the ancestors of modern-day humans, and that this is a possible rationale for differing frequencies of Neanderthal-specific DNA in the genomes of modern humans. Vernot and Akey (2015) concluded that the relatively greater quantity of Neanderthal-specific DNA in the genomes of individuals of East Asian descent than those of European descent cannot be explained by differences in selection.[21] They further suggest that "two additional demographic models, involving either a second pulse of Neanderthal gene flow into the ancestors of East Asians or a dilution of Neanderthal lineages in Europeans by admixture with an unknown ancestral population" are parsimonious with their data.[21] Similar conclusions were reached by Kim and Lohmueller (2015): "It has been hypothesized that the greater proportion of Neanderthal ancestry in East Asians than in Europeans is since purifying selection is less effective at removing weakly deleterious Neanderthal alleles from East Asian populations. Using simulations of a broad range of models of selection and demography, we have shown that this hypothesis cannot account for the higher proportion of Neanderthal ancestry in East Asians than in Europeans. Instead, more complex demographic scenarios, most likely involving multiple pulses of Neanderthal admixture, are required to explain the data."[22]
Khrameeva et al. (2014), a German-Russian-Chinese collaboration, compiled a consensus Neanderthal genome based on the genome of the Altai individual and of three Vindjia individuals. This was compared to a consensus chimpanzee genome as the outgroup and to the genome of eleven modern populations (three African, three East Asian, three European). Beyond confirming the significantly higher similarity to the Neanderthal genome in non-Africans than in Africans, the study also found a difference in the distribution of Neanderthal-derived sites between Europeans and East Asians, suggesting recent evolutionary pressures. Asian populations showed clustering in functional groups related to immune and hematopoietic pathways, while Europeans showed clustering in functional groups related to the lipid catabolic process. [23] Evidence for AMH admixture to Neanderthals at roughly 100,000 years ago was presented by Kuhlwilm et al. (2016). [24]
Figure 1: How Neanderthals affected gene flow in humans
There have been at least three episodes of interbreeding. The first would have occurred soon after some modern humans left Africa. The second would have occurred after the ancestral Melanesians had branched off—these people seem to have thereafter bred with Denisovans. The third would have involved Neanderthals and the ancestors of East Asians only. [25-27]
A 2016 study presented evidence that Neanderthal males might not have had viable male offspring with AMH females. This could explain why no modern man to date has been found with a Neanderthal Y chromosome. [28] A 2018 study concluded that interbreeding between Neanderthals and modern humans led initially to the exposure of each species to unfamiliar viruses. Later on, the exchange of genes granted resistance to those viruses, too. [29]
On July 3, 2020, scientists reported finding that a major genetic risk factor of the Covid-19 virus was inherited from archaic Neanderthals 60,000 years ago. [7][8] [30] It is estimated that 16% of Europeans and 50% of South Asians have the particular sequence on chromosome III, with 63% of Bangladeshis having these gene sequences. Africans, Middle Easterners, and East Asians reported the presence of the chromosome in very negligible amounts. [31-33]
If someone becomes infected with the coronavirus SARS-CoV-2, he/she might wish there was a fast way to check his/her Neanderthal ancestry. A small but significant number of people have an ancient gene variant from the extinct hominin that may double, or even quadruple, their risk of serious complications from COVID-19. The finding shines a light on an enzyme called dipeptidyl peptidase-4 (DPP4). Scientists already know the protein allows another coronavirus, which causes Middle Eastern respiratory syndrome (MERS), to bind to and enter human cells. The new analysis, of DPP4 gene variants among COVID-19 patients, suggests the enzyme also provides SARS-CoV-2 with a second door into our cells, along with its usual infection route via the angiotensin-converting enzyme 2 (ACE2) receptor on cell surfaces. [34-37]
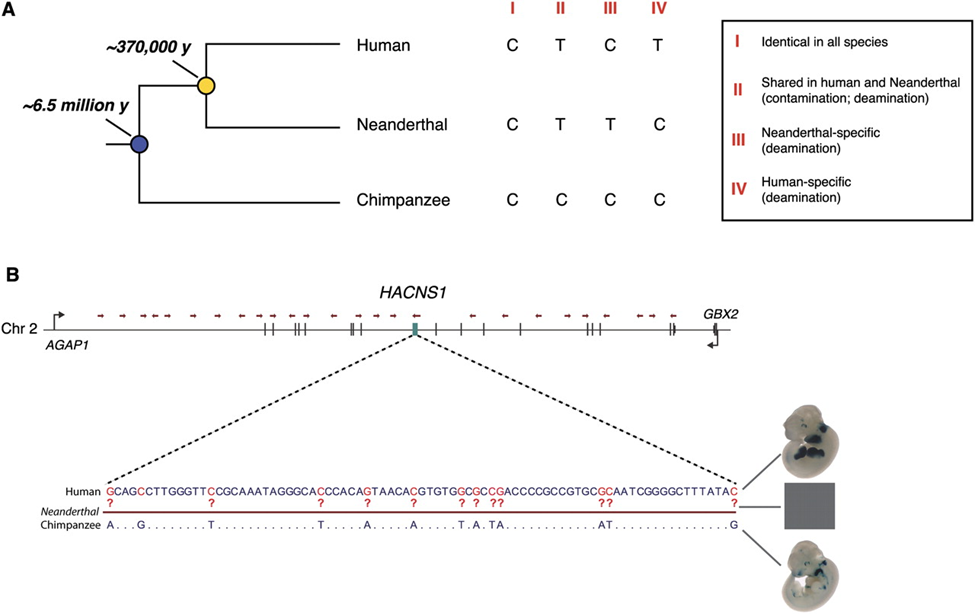
Figure 2: Evolutionary relationship of modern human, Neanderthal, and chimpanzee. (James P. Noonan, 2010)
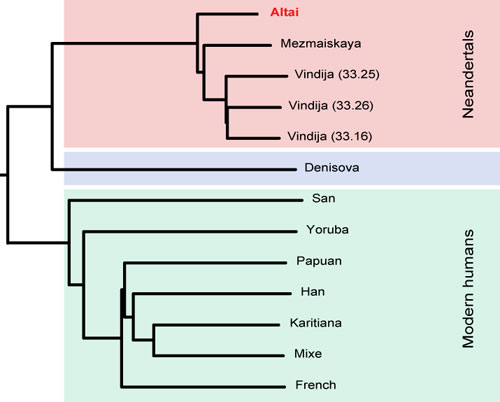
Figure 3: DNA sequences were generated on the Illumina HiSeq platform and constitute an average 50-fold coverage of the genome. 99.9% of the 1.7GB of uniquely mappable DNA sequences in the human genome are covered at least ten times
Contamination with modern human DNA, estimated from mitochondrial and nuclear DNA sequences, is around 1%. The figure shows a tree relating this genome to the genomes of Neanderthals from Croatia, from Germany and from the Caucasus as well as the Denisovan genome recovered from a finger bone excavated at Deniosva Cave. It shows that this individual is closely related to these other Neanderthals. Thus, both Neanderthals and Denisovans have inhabited this cave in southern Siberia. Other groups looking in genetic databases for factors that influence COVID-19 severity have not flagged the DPP4 gene. But the work is provocative because it suggests some diabetes drugs, which target the cell surface protein, could help treat the disease. We want to put this finding out there quickly so people can systematically test if DPP4 could be a therapeutic target in patients with Coronavirus. [38-39]
DPP4 may play a role in the infection of SARS-CoV-2. DPP4 should be a good binding partner for the protein called spike on the surface of the SARS-CoV-2 virus, based on comparing amino acid sequences and crystal structures of the enzyme and spike’s established partner, ACE2. Another team, however, had earlier ruled out DPP4 as a SARS-CoV-2 receptor after finding the virus did not bind with it in cell line studies. [40]
Pääbo and co-author Hugo Zeberg, also an evolutionary geneticist at Max Planck, have now highlighted DPP4 again. Most Europeans, Asians, and Native Americans harbor a handful of genes from Neanderthals, up 1.8% to 2.6% of their DNA.
Studies of ancient DNA in Neanderthal fossils have shown the hominin's DPP4 gene subtly differs from the typical human one. Pääbo and Zeberg examined whether that Neanderthal gene variant or others from the extinct species appear more often in people with severe cases of COVID-19 than in uninfected people. For that, they turned to the latest data released in October from the COVID-19 Host Genetics Initiative, which has collected genome information and COVID-19 status on many people from other studies or data banks. [41-46] They only searched for Neanderthal versions of genes in people who had had severe COVID-19, which gave them a quick way to see whether these archaic genes influenced how living people responded to the coronavirus. The Neanderthal version of DPP4 popped up at a higher frequency in the genomes of 7885 people hospitalized with severe COVID-19 than in a control group. If a person had a single copy of the Neanderthal gene variant, they had double the risk of severe COVID-19 when infected, if both their copies of DPP4 were Neanderthal, their risk quadrupled. [47]
The researchers estimate that between 1% and 4% of Europeans and Asians have inherited a Neanderthal version of the DPP4 gene. [48-50]
A 2018 study by David Enard found that living humans have inherited a disproportionate number of Neanderthal variants of immune genes that target RNA viruses like coronaviruses, compared with genes that respond to DNA viruses. [51-52]
Science’s COVID-19 reporting is supported by the Pulitzer Center and the Heising-Simons Foundation. It’s one of the pandemic’s puzzles: Most people infected by SARS-CoV-2 never feel sick, whereas others develop serious symptoms or even end up in an intensive care unit clinging to life. Age and preexisting conditions, such as obesity, account for much of the disparity. But geneticists have raced to see whether a person’s DNA also explains why some get hit hard by the coronavirus, and they have uncovered tantalizing leads. [53-55]
A U.K. group studying more than 2200 COVID-19 patients has pinned down common gene variants that are linked to the most severe cases of the disease, and that points to existing drugs that could be repurposed to help. Each one provides a potential target for treatment. [56-69]
Conclusion
The hominin's DPP4 gene subtly differs from the typical human one. Pääbo and Zeberg examined whether that Neanderthal gene variant or others from the extinct species appear more often in people with severe cases of COVID-19 than in uninfected people. For that, they turned to the latest data released in October 2020 from the COVID-19 Host Genetics Initiative, which has collected genome information and COVID-19 status on many people from other studies or data banks. They only searched for Neanderthal versions of genes in people who had had severe COVID-19, which gave them a quick way to see whether these archaic genes influenced how living people responded to the Coronavirus. If a person had a single copy of the Neanderthal gene variant, they had double the risk of severe COVID-19 when infected, if both their copies of DPP4 were Neanderthal, their risk quadrupled. The fact is Neanderthal Genes in modern humans, lowered the Immune System and faced them with many diseases such as HPV, Diabetes, Flu, COVID-19 and many infectious diseases. All humans contain this gene from 1 to 4 percent. East Asians, Europeans, Middle and south Americans conveys more, hence; native Africans contain less amounts of hominin's DPP4 gene. Therefore; East Asians, Europeans, Middle and south Americans are prone to severe COVID-19.