Clinical Case Reports and Clinical Study
OPEN ACCESS | Volume 12 - Issue 5 - 2025
ISSN No: 2766-8614 | Journal DOI: 10.61148/2766-8614/JCCRCS
Hanieh Faizmahdavi1 , Marjan Assefi2* , Sohila Nankali3 , Alireza Sharafshah4,5, Vahid Omarmeli4,6
1Department of Obstetrics and Gynecology, Clinical Research Development Center, Imam Reza hospital, Kermanshah University of Medical Sciences, Kermanshah, Iran.
2CEO of Marie Curie Science Research Center, USA
3North Central University, USA
4Dr. Shaveisi-zadeh Medical Genetic Lab, Kermanshah, Iran.
5Division of Genetics, Department of Cell and Molecular Biology and Microbiology, Faculty of Science and Biotechnology, University of Isfahan, Isfahan, IR Iran.
6Biology Department, College of Bioscience, Islamic Azad University, Tehran North Branch, Tehran, Iran.
*Corresponding Author: Marjan Assefi, CEO of Marie Curie Science Research Center, USA.
Received: February 04, 2023
Accepted: February08, 2023
Published: February 10, 2023
Citation: Hanieh Faizmahdavi, Marjan Assefi, Sohila Nankali, Alireza Sharafshah, Vahid Omarmeli (2023) “A novel mutation in VPS13B gene causing Cohen Syndrome and representing new symptoms found by Whole Exome Sequencing”, Clinical Case Reports and Clinical Study, 1(9); DOI: http;//doi.org/02.2023/1.161.
Copyright: © 2023 Marjan Assefi. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly Cited.
Former case reports have indicated the wide spectrum of Cohen syndrome (CS). This syndrome has severe impacts on neuronal, ocular, muscular, gastrointestinal, and cardiovascular systems. Whole Exome Sequencing (WES) was done for a 5-year-old boy following by screening of novel mutation in his parents through the Sanger sequencing technique. Clinical tests were included in detail and compared with the previous reports of CS signs. WES analysis revealed that a novel homozygote mutation of the VPS13B gene (exon45:c.T8228C:p.L2743P) in the case. Also, sequencing of his parents represented heterozygosity of the mutated allele for both of them. Altogether, our findings extended the formerly described phenotypes of CS in an Iranian 5-year-old boy including motor-sensory disabilities, facial features, ocular impairments, and speech difficulties. More importantly, we report that lung infections and just one kidney formation is part of the newly phenotypic spectrum of CS associated with novel mutation L2743P in the VPS13B gene.
Introduction
As an autosomal recessive syndrome, Cohen syndrome (CS) has clinical characteristics including facial features accompanied by motor neuron postponement in development. This syndrome involves systems including ocular, musculoskeletal, nervous, gastrointestinal, hematologic, cardiovascular, and endocrine (Dehghan, Behnam et al. 2021). Nonsense or missense mutations in the Vacuolar protein sorting 13 homolog B (VPS13B) are reported to be the cause of Cohen syndrome with over 150 known variants documented in the Human Gene Mutation Database(Koehler, Schuelke et al. 2020). Members of the VPS13 protein family are involved in membrane fusion processes and mechanisms of vesicular transportation. While mutations in VPS13B cause SC, VPS13A, and VPS13C genes cause autosomal recessive neurodegenerative Huntington's disease and Parkinson's-like syndrome, respectively (Momtazmanesh, Rayzan et al. 2020, Li, Bu et al. 2021).
VPS13B as a fundamental protein for keeping the integrity and function of the Golgi apparatus has a high number of residues (more than 4000 amino acids) with lipid binding capability. Probably, various protein-protein interactions inherent in the architectures of the VPS13 domain are the cause of various manifestations of human disease induced through the vps13 gene family (Seifert, Holder‐Espinasse et al. 2009, Rejeb, Jilani et al. 2017).
Here, for the first time in the literature, we report a 5-year-old boy having a new mutation in the VPS13B gene (p.L2743P) with homozygote type showing Cohen syndrome symptoms.
Case Report
A 5-year-old boy with symptoms of movement disorder and a history of eye problems and lung infection at birth was admitted to the genetic laboratory (Figure 1). His parents had a family history of marriage (cousin). His parents expressed that after the birth, the 7-day-old baby boy could not open his eyes easily and his movement symptoms became more and more severe day by day. Two-month, he was hospitalized due to a lung infection and could not stand on his own two feet (shrinking his legs). Unfortunately, he was not diagnosed with the disease until he was 2 months old. He had a lung infection again at six months. After seeing a pediatrician, it was found that his head circumference was less than normal. MRI showed that the cerebellum was smaller than normal. A healthy ear, brain, and retina were diagnosed. Then, at age 5, WES showed that he had a new mutation in the exon 45 of the VPS13B gene. After the patient was referred to the parents during the genetic counseling session (the case had 5 years old), certain physical and behavioral symptoms were identified, which included: stretched face and skull, drooping eyelids with no looking up, circular movements of the neck and head, outstretched hands, and abnormally clenched fingers, recurring restlessness and impatience, overreacted happiness leading to a heavy heartbeat, hyperactivity, signs of semi-aggressive behaviors (biting the sleeve with a tooth).

Figure 1. Front (A) and profile (B) images of a 5-year-old boy with Cohen syndrome symptoms.
This paper obtained the informed consent of all patients and is consisted with the declaration of Helsinki. Genomic DNA was fragmented and enriched for exome sequences using the SureSelect Human All Exon Kit version 6 and sequencing was done at a minimal average coverage of 90X on an Illumina HiSeq 4000 platform. The initial sequencing component of this test was performed by the Illumina Whole-Genome Sequencing Service in Macrogen (Seoul, Korea), and the alignment, variant calling, data filtering, and interpretation were accomplished. Briefly, reads were aligned to the human reference sequence (GRCh37) with the Burrows-Wheeler Aligner (BWA), and variant calls are made by the Genomic Analysis Tool Kit (GATK). Variants were filtered to recognize the most potential candidate variants. The evidence for phenotype-causality was then evaluated for every variant obtained from the filtering strategies. Only those variants with evidence for causing or contributing to the disease are reported. Each variant was evaluated based on the available information from the following: databases (including HGMD, ClinVar, LSDBs, NHLBI Exome Sequencing Project, 1000 Genomes, and dbSNP), published literature, clinical correlation, and its predicted functional or splicing impact using evolutionary conservation analysis and computational tools (including AlignGVGD, MAPP, MutationTaster, PolyPhen-2, SIFT, and SNAP).
Clinical characteristics of the case were completely investigated and all files were archived with the full consent of the patient's parents. Neonatal screening of blood conditions did not give any indication of congenital hypothyroidism (TSH level), congenital adrenal hyperplasia (17-OH-progesterone), galactosemia (Gal+ Gal 1-P, Gal-1P-uridyltransferase), profound biotinidase-deficiency (biotinidase). The patient was diagnosed with mild hearing loss during general neonatal screening. Ultrasound of the kidneys and bladder showed no clear tissue from the right kidney. The left kidney was 83 mm long with echo and normal parenchymal thickness without stones and hydronephrosis was seen. On the right side of the abdomen, in the vicinity of the sow muscle, a cyst containing echogenic debris with definite boundaries and a thin wall measuring 21 x 34 mm was seen, which looked like a mesenteric cyst. The bladder wall was slightly thick.
WES was done for the case and a novel variant was observed in exon 45 of the VPS13B gene. Due to the homozygosity in the case, Sanger sequencing was performed on the case’s parents by Sanger sequencing at the mutation site. Confirmation of mutation on variant revealed that mutation in VPS13B gene is new and parents are both heterozygotes (Figure 2). This mutation follows the autosomal recessive genetic inheritance. This mutation will cause Cohen syndrome, which is consistent with the clinical symptoms of the case.
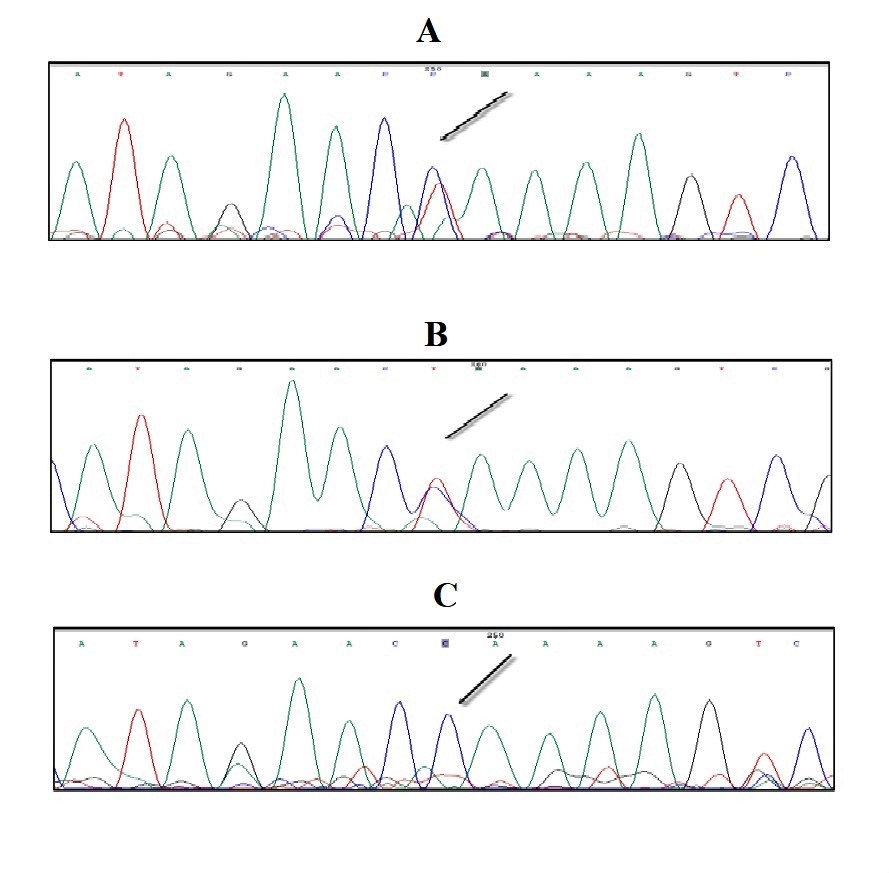
Figure 2. The sequence results of the case’s mother (A), his Father (B), and himself (C) represent heterozygote (TC), heterozygote (TC), and homozygote (CC) genotypes, respectively.
Discussion
In the current paper, we report a novel mutation in exon 45 of the VPS13B gene in an Iranian family by WES and Sanger sequencing tests. A 5-year-old boy born from parents both carrying the mutant allele as heterozygotes was introduced with Cohen syndrome signs including ocular impairment, movement disorders, certain facial symptoms, lung infection, hyperactivity, and the lack of kidney formation. Increasing reports of VPS13B novel mutations make CS a concerning genetic disorder that need to be screened in the prenatal diagnosis.
Previous studies reported that VPS13B has extensive-expression in the brain, blood, small intestine, muscles, placenta, heart, retina, kidney, and lung (Seifert, Holder‐Espinasse et al. 2009, Rejeb, Jilani et al. 2017). Rejeb et al found novel mutations in the VPS13B gene among Tunisians (Rejeb, Jilani et al. 2017). Koehler et al’s study documented a novel mutation of VPS13B in a German family (Koehler, Schuelke et al. 2020). Li et al’s study recently showed a splice-site mutation causing a skip of exon 38 in a Chinese family (Li, Bu et al. 2021). New reports by Momtazmanesh et al and Dehghan et al both from Iran (Momtazmanesh, Rayzan et al. 2020, Dehghan, Behnam et al. 2021) have raised concerns about the spread of new and unknown mutations causing this disease and emphasize prenatal genetic diagnosis.
Present study demonstrated the that the amino acid change form Leucine to Proline can have important impacts on the protein structure; So that Leucine is one of the amino acids with hydrophobic side chain and amino acid Proline, which changes the direction of the protein backbone and causes a turn in the secondary structure and finally the final structure of the protein. Therefore, it might be suggested that the amino acid change from Leucine to Proline can make a notable difference in protein folding. Also, because this mutation occurs approximately in the middle of the protein sequence, it may alter functionally sensitive regions.
In conclusion, the present study, in consistent with other recent studies related to reports of new mutations in the VPS13B gene, is adding new clinical features to CS. This has raised concerns about the prevalence of CS, and highlights the importance of genetic screening for the disease in prenatal testing.
Acknowledgement
We are thankful from all of our colleagues in the Dr. Shaveisi-zadeh lab for their sincere and compassionate cooperation.
Disclosure statement
No conflict of interest was reported by the authors.
Authors ‘contributions
Parichehr Darabi did the whole wet lab experiments. Dr. Masoumeh Favaedi contributed in the detection and diagnosis the disease and helped with the writing the draft of the paper. Dr. Nasrin Mansouri contributed in the data preparing and checking the results. Dr. Hanieh Faizmahdavi contributed in the consultation process and providing the patient profile. Dr. Soheila Nankali and Dr. Marjan Assefi contributed in the paper preparation. Dr. Alireza Sharafshah contributed in the bioinformatic analysis and writing the paper. Dr. Zhila Shaveisi-zadeh helped with the writing and revising the final edition of the paper. Finally, Dr. Vahid Omarmeli supervised the design, data preparation, and final editing of the paper.
Ethical approval and consent to participate
All the participants were able to give consent to participate in the study and signed a written consent form. The study was carried out in accordance with the Declaration of Helsinki.
Data availability statement
The data is not located on an open server, but could be made available on request to the corresponding author.